The growing catalogue of online biodiversity data sources has
reached a scale where data integration and mashups can be
truly useful. Information about species is being aggregated at
national and international scales. Here we provide a
visualization with template web applications, phyloJIVE, which
places biodiversity web-services into an evolutionary context.
You can download
files to build your own instance of phyloJIVE. It includes
template web application, instructions and other samples. The
template web application can be incorporated into a webserver
(as is) or can run by itself on a PC (windows users: extract
the folder on your desktop).
Further documentation, notes on common problems, and how to
tailor the configuration to your needs (and data sources) is
available on the phyloJIVE wiki
or on the getting started page
(if you are impatient). Notes on charJSON along with
scripts are available here.
Details on phyloJIVE web application parameters are here.
Further details at development site where
contributions and comments are encouraged.
Examples
- Acacia Case
Study: A phylogeny with some characters.
--> Demonstrates simple the algorithms for naive ancestral trait inference.
- Hornworts: A small phylogeny
of Australian Hornworts
--> including some geophysical characteristics for each species.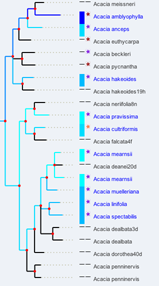
- Hornworts shows how to customize menu and hovers.
- Syncarida - shows how to use
LSID's
-->Taxon names are looked up in a nomenclator. The result is used to pull back a map keyed to the LSID for the taxon/species
--> demonstrates how to send a clade to the ALA Spatial portal. - Australian extent
endemic birds: A bird phylogeny which shows the
relationship of species endemic to Australia
-->(i.e. excluding Extinct/overseas species). - Amphibia. A moderate
size phylogeny (~2800) of extant frogs, salamanders, and
caecilians
-->linked only to the EOL and DiscoverLife.
-->Demonstrating performance. - Marsupials 1 - A
species Level phylogeny of extant marsupials
--> uses the template to demonstrate linking. - Marsupials 2 - A species
Level phylogeny of extant marsupials
--> demonstrates how to send a clade to the ALA Spatial portal.
- RFK Rainforest plants of Northern Australia - including characters
- A LARGE phylogeny - coming soon. Demonstrating performance.
integration with Atlas of
Living Australia and IdentifyLife
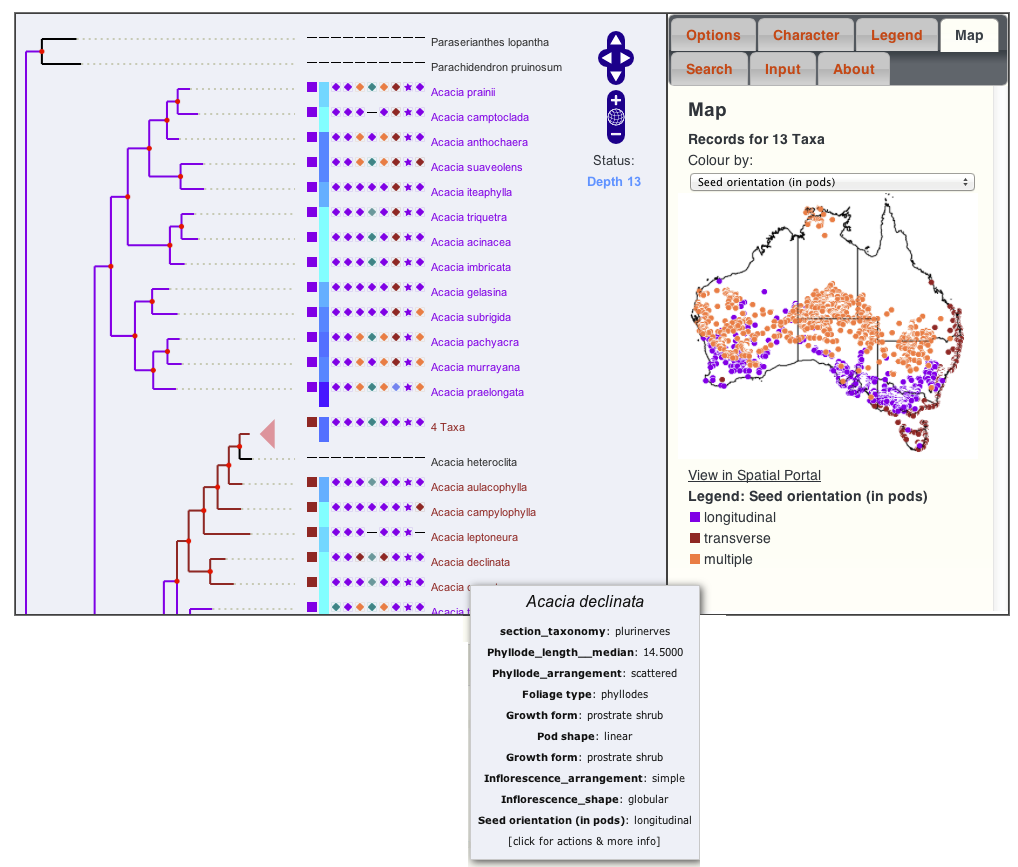
- Acacia Case Study: which can be linked to +180 characters from "The Key to Australian Acacia" by Bruce Maslin. Characters for clades can be mapped in the ALA spatial portal (of up to 32 taxa)
- Coral Case Study: of the impact of non monophyletic classification on conservation agendas.
- Marsupial
Case study: clades containing IUCN listed endangered
species can be placed into a biogeographical context via the
ALA spatial portal (e.g. the cuscoids)
about
phyloJIVE is a web based application that places biodiversity information aggregated from many sources onto compact phylogenetic trees. It is
- is entirely client-side
- renders in the web browser on a HTML 5 canvas.
- requires no plugins
- is open source
- works in current versions of all major browsers
- is built on the TheJit (javascript infovis library)
Joe Miller's has made detailed
instructions on the use the ALA implementation of phyloJIVE
including videos. We will also be adding details to the
project wiki.
There are of course, known
and unknown problems. You are definitely
encouraged to report any bugs in the project
wiki, notify us, or to fix them.
What can the end user do in the browser?
with the template* web
application the user can.....
- Change the order of branches and clades
- Ladderise
- Search for taxa
- Select up to three characters for simultaneous display on the tree
- Change scale
- Collapse / expand select clades
- Set new roots for the tree
- Explore taxon specific information pulled from pre-configured webservices
- Input their own (newick formatted) phylogentic tree for linkage to predetermined webservices including CHARJSON character services
* of course you are welcome to roll your own) you are welcome to roll your own webapp around the visualization as done by the Atlas of Living Australia
Integration
Web-services phyloJIVE can be configured easily to
asynchronously access information from many online
biodiversity data sources. Examples providers include The
Atlas of Living Australia, The Catalogue of Life, The National
Species List and The World Register of Marine Species. This
information is accessed and displayed as the tree is explored
by end users dynamically and immediately on hover, in summary
tables, and in detail.
Performance
Performance is determined by the viewer’s computer and
browser.
Trees can have at least 4000 taxa with matrices of at least
100 characters.
Characters (Species and Taxonomic)
Up to three characters from a matrix can be chosen for
comparison at a time.
- Putative differences in the ancestral states qualitative characters are inferred from (Fitch) parsimony.
- Putative changes in the ancestral states of quantitative characters are inferred from a phylogenetically weighted comparison of means and standard deviations.
- The reconstructed ancestral state of the first character is used to determine branch colour. Transitions in any of the three characters is indicated by a red box.